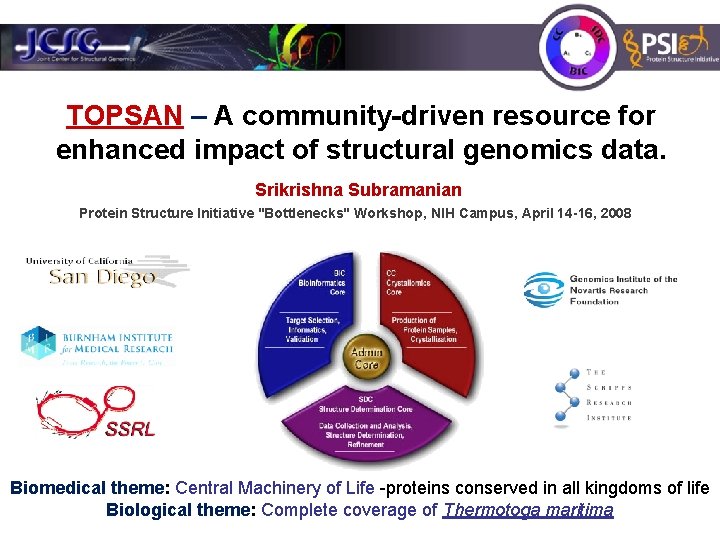
JCSG
JCSG database
- The ABM database links to other important information and it is easily add references, classification widgets for SCOP annotation.
- WYSIWYG editor, author and version tracking system (critical for developing publication authorship), tagging system, metabolic pathway information for Thermotoga maritima proteins and human proteomics.
There are no products listed under this category.
